Biofilm Integrated Nanofiber Display (BIND)
This is a central platform technology developed by our lab. We seek to continue its development as a versatile and practical materials synthesis method using a combination of biopolymer and organismal engineering.
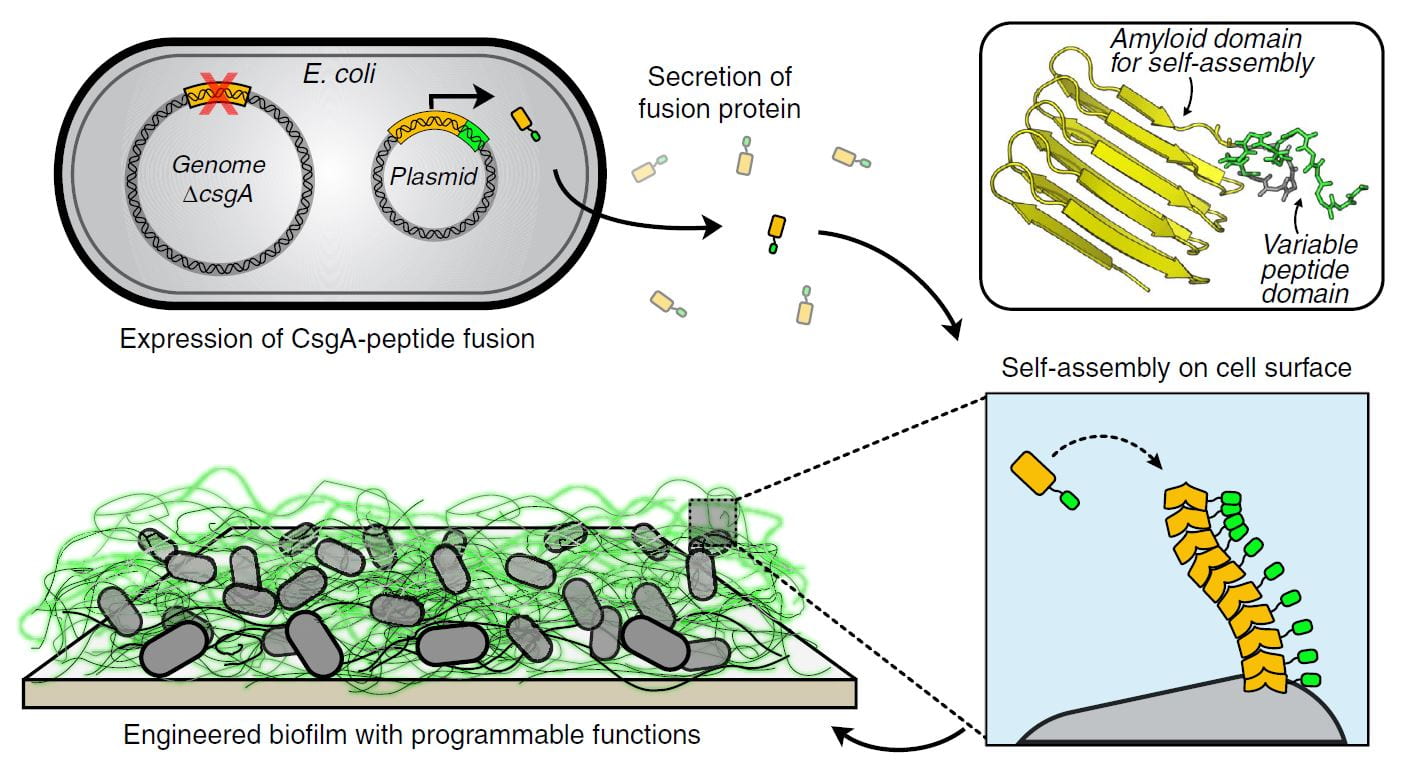
Biofilm Integrated Nanofiber Display repurposes matrix proteins normally produced by E. coli during biofilm formation as a platform for biopolymer engineering. The technology focuses on the curli system of E. coli, but could be ported to other organismal chassis. Curli are functional amyloids composed primarily of a single protein monomer (CsgA). CsgA is secreted into the extracellular space, where it is nucleated by cell surface proteins to form a beta-helical fold. Additional monomers are added to form fibers many times the cell length. BIND uses the machinery that already exists in E. coli, dedicated to CsgA secretion, to produce fibers composed of engineered CsgA variants. The resulting fibers can be imbued with a range of functional properties, for example the display of specific binding domains, crosslinking domains, or enzymes. In turn, the materials made from these fibers exhibit properties – like specific surface adhesion, molecular recognition, or catalysis – that would be difficult to obtain with conventional synthetic polymers. Ongoing projects in the group are focused on engineering strains with enhanced secretion capabilities and the ability to secrete and assemble increasingly complex biopolymer architectures.
A paper describing the use of Congo Red dye fluorescence to monitor curli fiber formation in growing bacterial cultures:
Kan, A., Birnbaum, D. P., Praveschotinunt, P., & Joshi, N. S. (2019). Congo Red fluorescence for rapid in situ characterization of synthetic curli systems. Applied and environmental microbiology, 85(13), e00434-19.
A paper describing the foundational exploration of BIND:
Nguyen, P. Q., Botyanszki, Z., Tay, P. K. R., & Joshi, N. S. (2014). Programmable biofilm-based materials from engineered curli nanofibres. Nature communications, 5(1), 1-10.